Introduction
We have a whole range of tools available here, including APIs, an index of species popularity and source code, but we're always working on the next enhancements and are trying to build a developer community. Please don't struggle on if you hit difficulties or have concerns, instead contact us so we can work together on solving your problem. If you've done something special with OneZoom please let us know. Perhaps you've used it for teaching, visualising your own data or in a public display? OneZoom is a non-profit organisation and our purpose is to advance the education of the public in the subjects of evolution, biodiversity and conservation of the variety of life on earth.
- Read the license, plain language license FAQs and readme documentation, ask us about any queries or concerns.
- Pull the latest version of OneZoom from our public GitHub repository and follow the instructions in the readme file.
- Contact us for a database dump (which will include everything we have except for sponsorship information). Alternatively run our Python scripts to generate your own data, but this will be harder and involves many steps including getting large source data dumps from Open Tree of Life and Wikipedia as well as intensive use of the IUCN and EOL APIs to acquire further data.
- Optional: get copies of the images from us so they can be hosted in an environment disconnected from the internet, such as a touch table without a network connection.
- Safari, Chrome, Mobile devices: full compatibility but please keep up your software up to date.
- Firefox: full compatibility but not as responsive as Safari and Chrome.
- Opera, EDGE, Internet Explorer 9 and above: near full compatibility occasionally minor features don't work.
- General: make sure that Javascript is enabled and that your browser supports HTML5 and canvas.
Hosting your own variation of the OneZoom complete tree of life explorer.
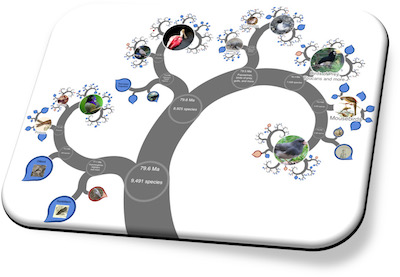
If you contact us we can provide help and add you to our developer community from which you can receive support. The keys steps to getting started are as follows...
Languages and frameworks used for OneZoom
The website is written as an application of the Web2py framework with SQL database. Most of the server side code is therefore written in Python, including those which are run outside of Web2py for data synthesis and preparation. The tree explorer itself is written in ECMAScript, which is compiled into javascript. The tree view is painted onto a javascript canvas and styled by functions wrapped inside the ECMAScript. All other pages, including the tree of life explorer user interface around the tree view, styled using SASS, which is compiled into CSS. We use Grunt and NodeJS for scripting compilation tasks. We have expanded on all this in a recent publication: "Dynamic visualisation of million-tip trees: the OneZoom project" Yan Wong, James Rosindell (2020)bioRxiv 2020.10.14.323055 doi: 10.1101/2020.10.14.323055. There is also extensive documentation in the readme files on our Github repository
OneZoom module structure
The OneZoom core is separated into three layers: external data acquisition, server-side synthesis of data, and the client-side tree of life explorer itself. Around the core there are two other components: our bespoke crowd funding system for managing leaf sponsorship (which is our primary means of funding) and other web pages outside of the tree viewers.
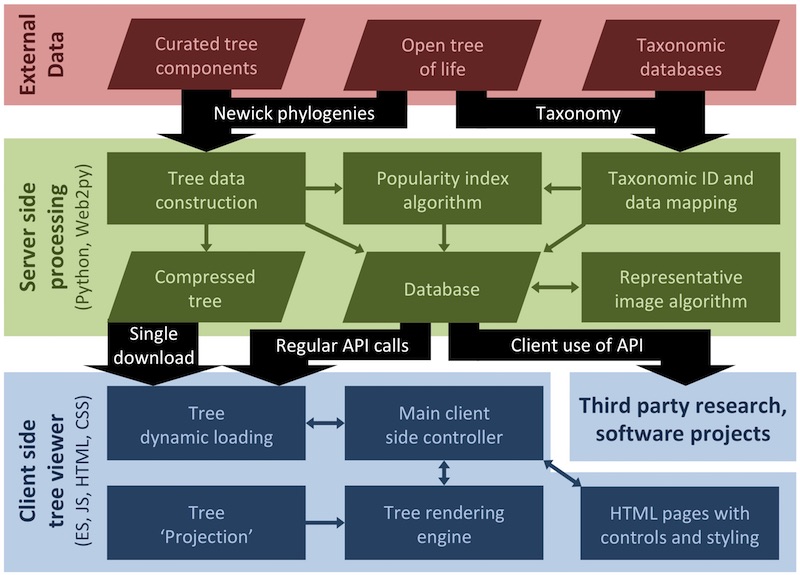
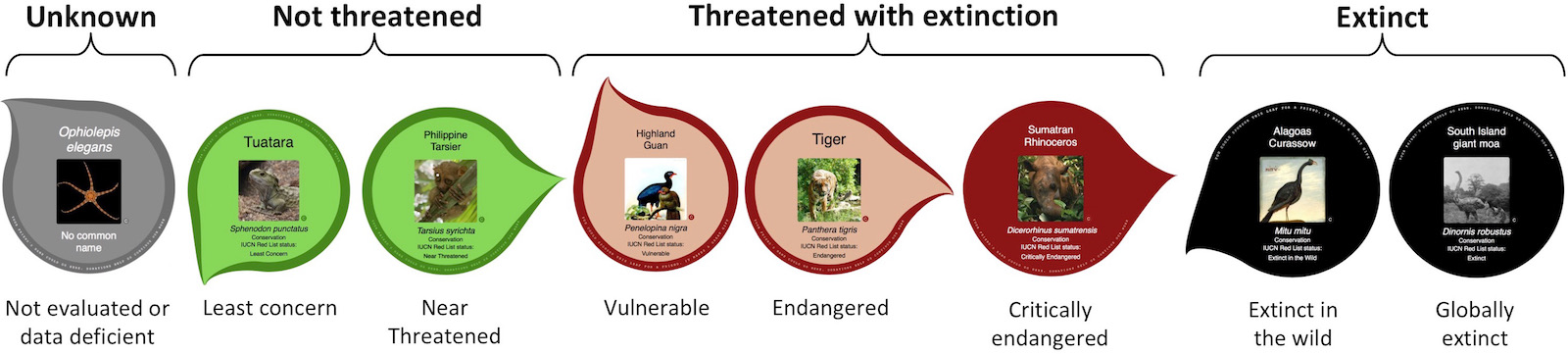
The database output includes the complete tree of life (mostly provided by the open tree of life) mapped with i) vernacular names, ii) extinction risk data from the IUCN, iii) IDs for species in a range of other resources, iv) our popularity index scores for all species and iv) representative images for interior nodes of the tree. We can imagine these data being useful for a wide range of third party projects not necessarily concerned with tree visualisation at all. For example Malachi Willey recently used the OneZoom back end to support the website Phyloquiz, a fun generator for phylogenetic quizzes that automatically generates multiple choice questions about the relationship between species. Another example is Phylotastic, which uses our popularity index as a way to automatically subset trees.
For projects focused more on visualisation of large trees within a zooming user interface, almost all of our codebase can be reused. The 'Tree Projection' module in our block diagram above defines a collection of lines and points to describe a large tree. The fact that it is abstracted from the rest of the code is important because it means that if you have a different way in mind to lay out a large tree, you can implement this whilst only making changes to the tree layout module. Your view would automatically benefit from access to the complete tree data, access to associated metadata, a zooming user interface, procedures to fly and jump around the tree, and all the performance optimisations that go on behind the scenes to much such large visualisations tractable.
The way in which leaves, nodes and branches are drawn at their defined points is defined by a separate module. So you could redesign the shapes of leaves and nodes, or the content that is displayed on them at different levels of zoom, and then reuse these leaves on any tree projection. The colour scheme of the tree, as a function of tree properties and metadata, is another abstracted block of code defining leaf, node and branch colours independently from their shapes or layouts. The rendering of shapes on canvas too is done in a separate module and could (in theory and with a bit of work) be swapped out for another rendering solution (e.g. WebGL or d3) without affecting the other parts of code. So, you could combine together any colour scheme, any leaf and node design, any tree 'projection' and any drawing module.
Web browser compatibility
Using source code from our public GitHub repository or Docker image
All our live source code is available to view on our public GitHub repository. We also provide two Docker images of our latest codebase: oztree-complete contains everything you need to run OneZoom with no live internet connection, oztree is a download of reduced size that uses the internet to grab images as needed from the OneZoom server.
Some parts of our codebase are available for reuse under OSI approved fully open source licenses. All other parts can be freely reused under a OneZoom non-profit source available license. This basically means they are free to use for the advancement of science and science education, but not for commercial purposes. We know that 'commercial' can be an elastic term, we are willing to interpret this in a relaxed way to support any project whose aims are consistent with our own mission statement. If you think this applies to you, please write to us and we'd be happy to consider providing you with a letter confirming that we don't consider your project to be commercial. For further details please see our plain language license FAQs.
Our objective is to encourage broad reuse of the OneZoom code whilst also protecting our unique model for maintaining the codebase and sustaining the live website. If the OneZoom organisation ceases to exist our software will automatically become available under an OSI approved MIT license. Remember that OneZoom is a registered not for profit organisation and the majority of work is done by unpaid volunteers. If you have suitable software development expertise and want to contribute please let us know, we'd be delighted to work with you.